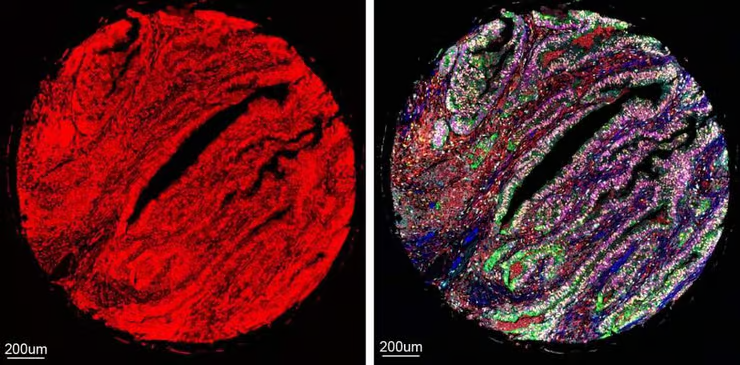
An illegible microscopy image with overlapping fluorescent labels of seven proteins (left) was translated with great precision using CombPlex into an image that allows the different proteins to be distinguished (right)
Artificial intelligence (AI) is transforming research by helping scientists understand complex biological data. A recent study from the Weizmann Institute of Science, published in the scientific journal Nature Biotechnology, showcases a groundbreaking AI-powered technology called CombPlex. Developed by Dr. Leeat Keren's team in the Department of Molecular Cell Biology, CombPlex allows researchers to observe more proteins in tissue samples than ever before, marking a significant advancement in studying cellular composition.

Why is this Important?
Dr. Keren explains that to understand how tissues function, it's crucial to measure as many proteins as possible concurrently. This is vital for a deeper knowledge of diseases like cancer, where knowing the makeup of tumor tissues can influence treatment effectiveness and predict patient recovery chances. However, traditional methods can only analyze three or four proteins at a time. CombPlex can measure around 20 proteins simultaneously in a single cell, with the potential to increase this number to hundreds in the future. This technology uses existing lab equipment, making it accessible for widespread use.
How Does CombPlex Work?
CombPlex uses fluorescent tags to label proteins, creating unique color combinations (barcodes) for each protein. AI algorithms then decode these barcodes, even when they appear as a tangled mess to the human eye. This method allows researchers to study many proteins quickly and accurately.
Impact and Future Prospects
CombPlex could revolutionize tissue research in labs and hospitals, providing faster and more comprehensive insights into tissue composition. Dr. Keren hopes this technology will replace current methods, offering deeper clinical insights and advancing medical research.
A version of this article was first published in ynetnews.


